|
Department of Botany, The Field Museum, Chicago, IL 60605 USA |
Department of Botany, The Field Museum, Chicago, IL 60605 USA |
Department of Botany, Charles Darwin Research Station, Isla Santa Cruz Galapagos, Ecuador |
Introduction
|
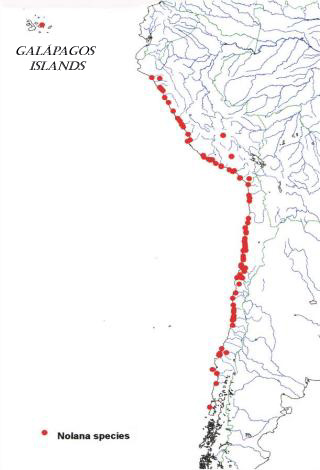
The vegetation of coastal Peru and northern Chile, termed "lomas" formations, is composed of floristic elements from various biogeographic sources (Dillon, 1997; Dillon & Hoffmann, 1997). With over 85 species, Nolana L. (Solanaceae-Nolaneae) stands out as the most wide-ranging and conspicuous floristic element of these formations (Fig. 1). The evolutionary history of Nolana may hold clues to the age and origin of the coastal deserts. In most modern classifications, members of Nolana have been recognized at the familial (Nolanaceae) or subfamilial (Nolanoideae) rank due to its unusual carpel morphology, but chloroplast DNA restriction site mapping studies have provided evidence that Nolana clusters within the family Solanaceae with closest relationships to Lycium L. and Grabowskia Schlecht. (Olmstead et al., 1999). Initial morphological and molecular systematic studies within Nolana have allowed the establishment of a putative phylogeny for the Nolana that provides a framework for testing hypotheses of character evolution and biogeographic relationships (Tago, 1999; Tago & Dillon, 1999). |
| Most species of Nolana are narrow endemics, with small, restricted geographic ranges and specific ecological requirements. Only one species, Nolana galapagensis (Christoph.) I.M.Johnst., has a distribution outside of continental South America. N. galapagensis is recorded from sandy dunes (Fig. 2) near the ocean on six islands within the Galápagos Island chain: Isabela, San Cristobal, Santa Cruz, Santa María, Seymour, and Tortuga (Wiggins & Porter, 1971; Eliasson, 1970; A. Tye, pers. comm.). The overall morphology of N. galapagensis is similar to northern Chilean species, specifically, N. sedifolia Poepp. (Fig. 3), (Fig. 4), sharing erect shrubby habit, reduced and succulent leaves, and small, white corollas. The latter species does not inhabit near-shore habitats, rather it is found in quebradas usually above 150 m. |
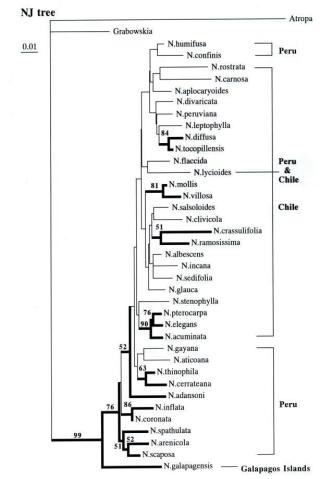
| Using a subset of 37 Nolana species, ITS sequence data was utilized in the reconstruction of a putative Nolana phylogeny (Tago, 1999). Parsimony analysis yielded strict consensus trees with incomplete resolution ("polytomy") , but placed N. galapagensis at the base of the clade, as the sister taxon to the remainder of the genus, i.e., all South American continental species. The grouping of continental species in a single clade is supported by bootstrap frequency of 76%. The phylogram resulting from neighbor-joining analysis of ITS sequence data (Fig. 5) also places N. galapagensis at the base and the next clade is composed of southern Peruvian species. While N. galapagensis shares greatest morphological similarity with N. sedifolia of northern Chile, its position on the ITS tree is distant from that Chilean species. (Fig. 5). This represents a paradox, given the overall morphological similarity to N. sedifolia, which is within a clade of other, small, white-flowered Chilean species, N. albescens and N. incana. The results of the preliminary ITS and matK studies have raised some intriguing questions, especially the ultimate position of N. galapagensis. |
| Our preliminary divergence
data allows the calculation of a range of potential dates for the arrival
of Nolana within the Galapagos Islands. In the ITS analysis, divergence
values for N. galapagensis approach those of the outgroup taxon,
Grabowskia. This high level of sequence divergence may be the product
of long-term isolation on islands from continental relatives or some other
phenomenon such as founder effect or genetic drift. Reported values suggest
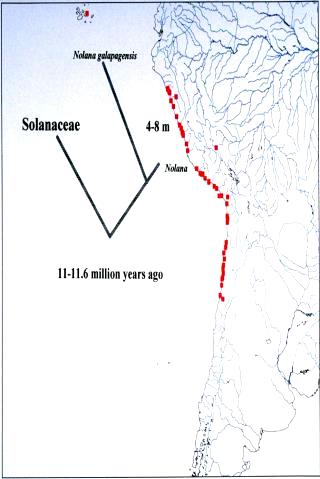
an average rate for ITS calculated at 5.20 x 10-9 per site per year (Tago,
1999). If this rate is adopted for Nolana, the divergence time of
Nolana from the closest taxon in the Solanaceae is estimated to be
11.6 million years ago (Fig. 6). Further, ITS sequence data suggests that
the divergence time of N. galapagensis from the rest of Nolana
species is ca. 8.1 million years ago. In comparison, the nucleotide substitution
in the coding region of the matK gene provided an average substitution
rate of 4.10 x 10-10, with the divergence time of Nolana from the
closest taxon in the Solanaceae is estimated at 11.0 million years ago.
With matK sequence data, the divergence time of N. galapagensis
from the rest of Nolana is estimated to be 4.0 million years ago.
Geological evidence points to an age for the current Galápagos archipelago
at 4-5 million years, but underwater seamounts may date to 15-20 million
years (Grehan, 2001). Molecular clock calibrations for determining colonization
events may provide estimates that exceed the age of the modern islands.
Until recently, very few detailed molecular studies had been conducted upon
the indigenous Galápagos flora (Elisens, 1992). As part of a broader study
of the evolutionary and biogeographic relationships within Nolana,
we propose to investigate N. galapagensis at the population level
with molecular systematics techniques. This approach will provide information
on the degree of genetic divergence between the various island populations
and will help to elucidate the age and origin of this unusual Galápagos
endemic. Knowledge of the amount of variability present within and between
populations will provide valuable information for addressing possible colonization
patterns and conservation priorities.
|
|
Materials and Methods To accomplish a population level study, not less than 10 individuals from each island will be sampled. A small amount of leaf material (i.e., 50 mg or ca. 20 leaves) will be gathered from each of the 10 individuals, with no harm to the plants. This material will be placed in plastic bags with an appropriate amount of silica gel to allow for fast drying. A limited number of voucher collections will be made and deposited in Charles Darwin Research Station herbarium (CDS). In some instances, small amounts of dried leaf material may be collected from herbarium sheets housed at CDS. In addition, samples of Grabowskia boerhaaviaefolia (L.f.) Schlecht and Lycium minimum C.L. Hitchc. will be gathered to provide outgroup taxa for the analysis. Grabowskia boerhaaviaefolia is recorded from seven islands: Champion, Española, Gardner, Las Plazas, Santa Cruz, Santa Fé, and Seymour. Its distribution on mainland South America includes Ecuador, Peru and Bolivia. Lycium minimum is a Galápagos endemic, recorded from nine islands, including Baltra, Español, Gardner, Isabela, Pinta, Pinzón, Santa Cruz, Santa Fé, and Santa María. It has been suggested that L. minimum is most closely related to L. californicum (Wiggins & Porter, 1971). Galápagos Island samples of the outgroup species will allow for comparisons with conspecific or congeneric continental populations in these genera. Total DNA will be extracted from silica gel dried leaf material. DNA amplifications will follow the protocol of Wen and Zimmer (1996) and the sequencing strategy of the entire ITS regions and the 5.8S gene will follow Wen et al. (2001). All newly-derived sequences will be deposited in GenBank. Phylogenetic analyses will be performed with PAUP* using maximum parsimony, maximum likelihood, and neighbor-joining methods. Populational analysis of Nolana galapagensis will be conducted with the amplified fragment length polymorphism (AFLP) techniques (Vos et al., 1995). This technique has been successfully employed to assess genetic diversity within and among populations of endangered plants (e.g., Travis et al., 1996). We will thus use this approach to examine the genetic structure of N. galapagensis and its conservation implication. |
|
Results The proposed research is designed to investigate a range of topics relating to the taxonomic status of Nolana galapagensis and the two outgroup species, Grabowskia boerhaaviaefolia and Lycium minimum. It is anticipated that the planned molecular systematic studies, as designed and using appropriate samples, will address the following evolutionary and biogeographic questions:
|
|
Literature CitedDillon, M.O. 1997. Lomas Formations-Peru, pp. 519-527. In: S. D Davis, V. H. Heywood, O. Herrera-McBryde, J. Villa-Lobos and A. C. Hamilton (eds.), Centres of Plant Diversity, A Guide and Strategy for their Conservation. WWF, Information Press, Oxford, U.K. [URL: http://www.nmnh.si.edu/botany/projects/cpd/sa/sa42.htm] Dillon, M.O., & A. E. Hoffmann-J. 1997. Lomas Formations of the Atacama Desert, Northern Chile, pp. 528-535. In: S. D Davis, V. H. Heywood, O. Herrera-McBryde, J. Villa-Lobos and A.C. Hamilton (eds.), Centres of Plant Diversity, A Guide and Strategy for their Conservation. WWF, Information Press, Oxford, U.K. [URL: http://www.nmnh.si.edu/botany/projects/cpd/sa/sa43.htm] Eliasson, U. H. 1970. Studies in Galápagos Plants IX, New Taxonomic and Distributional Records. Botaniska Notiser 123: 346-357. Elisens, W. J. 1992. Genetic Divergence in Galvezia (Scrophulariaceae): Evolutionary and Biogeographic Relationships among South American and Galápagos Species. Amer. J. Bot. 79: 198-206. Grehan, J.R. 2001. Biogeography and evolution of the Galapagos: integration of the biological and geological evidence. Biol. Journ. Linn. Soc. 74: 267-287. Olmstead, R.G., J.A. Sweere, R.E. Spangler, L. Bohs, & J.D. Palmer. 1999. Phylogeny and provisional classification of the Solanaceae based on chloroplast DNA. Pp. 111--137. In: M. Nee, D.E. Symon, R.N. Lester, & J.P. Jessop (eds.) Solanaceae IV: advances in biology and utilization. Royal Botanic Gardens, Kew. Tago, M. 1999. The Evolution of Nolana L. (Solanaceae) at lomas in South America. PhD. dissertation. Tokyo Metropolitan University. Tago-Nakawaza, M. & M.O. Dillon. 1999. Biogeografía y Evolución en el Clado Nolana (Solaneae-Solanaceae). Arnaldoa 6(2): 81-116. 1999 (2000). Travis, S.E., J. Maschinski, & P. Keim. 1996. An analysis of genetic variation in Astragalus cremnophylax var. cremnophylax, a critically endangered plant, using AFLP markers. Molecular Ecology 5: 735-745. Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. Van de Lee, M. Hornes, A. Frigiters, J. Pot, J. Peleman, M. Kuiper & M. Zabeau. 1995. AFLP: a new technique for DNA fingerprinting. Nucl. Acids Res. 23: 4407-4414. Wen, J., G.M. Plunkett, A.D. Mitchell, & S.J. Wagstaff. 2001. The evolution of Araliaceae: A phylogenetic analysis based on ITS sequences of Nuclear Ribosomal DNA. Syst. Bot. 26: 144-167. Wen, J. & E.A. Zimmer. 1996. Phylogeny and biogeography of Panax L. (Araliaceae): Inferences from ITS sequences of nuclear ribosomal DNA. Molecular Phylogenetics and Evolution 6: 167-177. Wiggins, I. L. & D. M. Porter. 1971. Flora of the Galápagos Islands. Pp. 1-998. Stanford University Press, Stanford, California.
Click on images for enlargements |